Division of Gene Expression Mechanism[Closed]
Link
・Division of Gene Expression Mechanism HP : http://www.www.fujita-hu.ac.jp/~mayeda/index-e.html
Research Interests
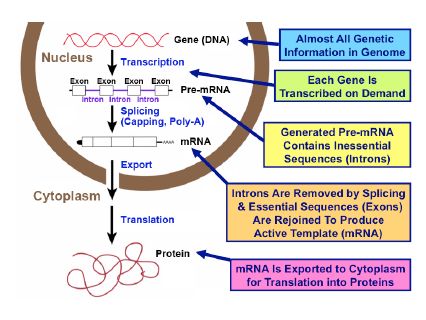
The cell nucleus, the central unit of living matter, contains genes (as DNA) that carry almost all genetic information. How is such important information processed and transferred to make the proteins essential for life?
The genes demanded by each living cell are first transcribed as mRNA precursors, or ‘pre-mRNAs’, and then all the inessential internal sequences are precisely removed to produce mature mRNAs (see right figure). This process, termed ‘splicing’, must be rigorously performed, or the mRNAs cannot act as correct templates for functional proteins. When this process is disturbed or dysregulated, important cell functions are disrupted, often with severe clinical consequences.
Our team in the Division of Gene Expression Mechanism has undertaken to clarify the unsolved mechanisms of this essential splicing to advance our basic understanding of the aberrant splicing that causes serious disease.
The genes demanded by each living cell are first transcribed as mRNA precursors, or ‘pre-mRNAs’, and then all the inessential internal sequences are precisely removed to produce mature mRNAs (see right figure). This process, termed ‘splicing’, must be rigorously performed, or the mRNAs cannot act as correct templates for functional proteins. When this process is disturbed or dysregulated, important cell functions are disrupted, often with severe clinical consequences.
Our team in the Division of Gene Expression Mechanism has undertaken to clarify the unsolved mechanisms of this essential splicing to advance our basic understanding of the aberrant splicing that causes serious disease.
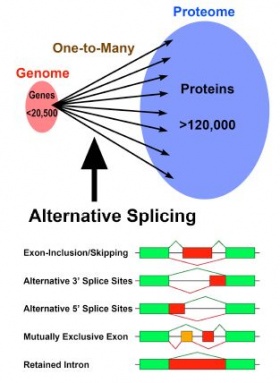
As higher eukaryotes have evolved, they have gained more and larger introns. In humans for instance, over 99% of the genomic sequence is introns, and consequently they massively interrupt genetic information on the genes. Introns vary extensively in length, from fewer than 40 to more than 1,000,000 nucleotides (> 25,000-fold difference). In fact, this variety of introns is precisely removed by splicing, which is apparently inefficient but extremely significant.
Alternative splicing allows the generation of multiple mRNAs from a single gene by changing the way of the pre-mRNA splicing, and the number of combinatorial patterns of splicing increases tremendously as the number of introns increases. In humans, alternative splicing is a successful major strategy for expressing the full proteome, with at least 120,000 kinds of proteins expressed from an unexpectedly small genome containing 20,500 genes at most (see left figure). Recent studies have revealed that over 90% of human genes undergo alternative splicing, which is spatiotemporally regulated on demand.
We study the basic mechanisms and functions in the gene expression process, which are essential for life, but they are also closely involved in human diseases and cancers. History demonstrates that basic studies can uncover the causes of diseases and provide the principles of radical therapies. We are studying hard in a small laboratory, but pursuing big ideas.
Alternative splicing allows the generation of multiple mRNAs from a single gene by changing the way of the pre-mRNA splicing, and the number of combinatorial patterns of splicing increases tremendously as the number of introns increases. In humans, alternative splicing is a successful major strategy for expressing the full proteome, with at least 120,000 kinds of proteins expressed from an unexpectedly small genome containing 20,500 genes at most (see left figure). Recent studies have revealed that over 90% of human genes undergo alternative splicing, which is spatiotemporally regulated on demand.
We study the basic mechanisms and functions in the gene expression process, which are essential for life, but they are also closely involved in human diseases and cancers. History demonstrates that basic studies can uncover the causes of diseases and provide the principles of radical therapies. We are studying hard in a small laboratory, but pursuing big ideas.
Selected Publications
1.T. Kameyama, H. Suzuku & A. Mayeda (2012). Re-splicing of mature mRNA in cancer cells promotes activation of distant weak alternative splice sites. Nucleic Acids Res. in press.
2.K. Ohe & A. Mayeda (2010). HMGA1a trapping of U1 snRNP at an authetic 5' splice site induces aberrant exon skipping in sporadic Alzheimer's disease. Mol. Cell. Biol. 30, 2220-2228.
3.J.H. Trembley, S. Tatsumi, E. Sakashita, P. Loyer, C.A. Slaughter, H. Suzuki, H. Endo, V. Kidd & A. Mayeda (2005). Activation of pre-mRNA splicing by human RNPS1 is regulated by CK2 phosphorylation. Mol. Cell. Biol. 25, 1446-1457.
4.E. Sakashita, S. Tatsumi, D. Werner, H. Endo & A. Mayeda (2004). Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Mol. Cell. Biol. 24, 1174-1187.
5.T. Manabe, T. Katayama, N. Sato, F. Gomi, J. Hitomi, T. Yanagida, T. Kudo, A. Honda, Y. Mori, S. Matsuzaki, K. Imaizumi, A. Mayeda & M. Tohyama (2003). Induced HMGA1a expression causes aberrant splicing of presenilin-2 pre-mRNA in sporadic Alzheimer's disease. Cell Death Differ. 10, 698-708.
6.A. Mayeda, J. Badolato, R. Kyobayashi, M.Q. Zhang, E.M. Gardiner & A.R. Krainer (1999). Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J. 18, 4560-4570.
7.A. Mayeda, S.H. Munroe, J.F. Caceres & A.R. Krainer (1994). Function of conserved domains of hnRNP A1 and other hnRNP A/B group proteins. EMBO J. 13, 5483-5495.
8.A. Mayeda & A.R. Krainer (1992). Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68, 365-375.
2.K. Ohe & A. Mayeda (2010). HMGA1a trapping of U1 snRNP at an authetic 5' splice site induces aberrant exon skipping in sporadic Alzheimer's disease. Mol. Cell. Biol. 30, 2220-2228.
3.J.H. Trembley, S. Tatsumi, E. Sakashita, P. Loyer, C.A. Slaughter, H. Suzuki, H. Endo, V. Kidd & A. Mayeda (2005). Activation of pre-mRNA splicing by human RNPS1 is regulated by CK2 phosphorylation. Mol. Cell. Biol. 25, 1446-1457.
4.E. Sakashita, S. Tatsumi, D. Werner, H. Endo & A. Mayeda (2004). Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Mol. Cell. Biol. 24, 1174-1187.
5.T. Manabe, T. Katayama, N. Sato, F. Gomi, J. Hitomi, T. Yanagida, T. Kudo, A. Honda, Y. Mori, S. Matsuzaki, K. Imaizumi, A. Mayeda & M. Tohyama (2003). Induced HMGA1a expression causes aberrant splicing of presenilin-2 pre-mRNA in sporadic Alzheimer's disease. Cell Death Differ. 10, 698-708.
6.A. Mayeda, J. Badolato, R. Kyobayashi, M.Q. Zhang, E.M. Gardiner & A.R. Krainer (1999). Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J. 18, 4560-4570.
7.A. Mayeda, S.H. Munroe, J.F. Caceres & A.R. Krainer (1994). Function of conserved domains of hnRNP A1 and other hnRNP A/B group proteins. EMBO J. 13, 5483-5495.
8.A. Mayeda & A.R. Krainer (1992). Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68, 365-375.
